A guide for students
What is Bioinformatics?
Bioinformatics brings together sub-areas of biology, information technology, computer science, mathematics, and statistics. The main objective is to conduct research and discoveries in biology, using expertise from other areas as a means to achieve this goal. For example, nowadays all areas of knowledge make use of the information technology (IT). So why do we need a specific discipline called Bioinformatics? The answer is based on two important characteristics of modern biological research. First, biology produces a lot of specific data. Second, we rely on the development of specific methods and algorithms to manipulate and analyse these data. Figure 1 shows possible relationships between areas of knowledge that contribute to researches in bioinformatics.
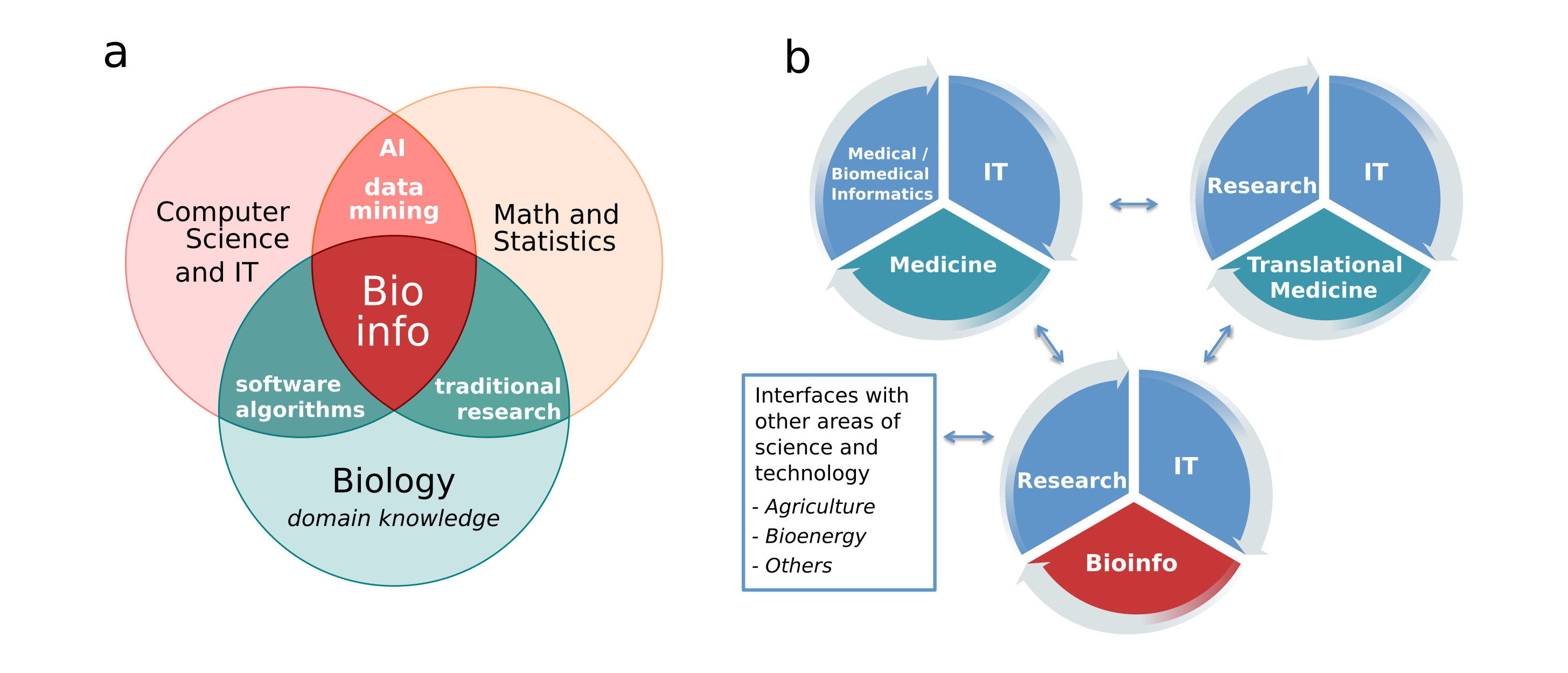
The main use of bioinformatics is to analyse data produced by molecular biology experiments. In its early days, molecular biology techniques were used to analyse individual genes, but now we can observe whole cells (for example, all genes at the same time) or even a community in a specific environment (for example, human intestinal microbiome). Many molecular biology techniques produce a large amount of data, such as DNA and protein sequences, coordinates of atoms in a protein structure, genes or proteins expressed in a cell at a given period of time and/or condition. All of these data should be stored and analysed, producing more information, which in turn should be stored and analysed with more information from other sources, and so on.
What does a Bioinformatician do?
According to Welch et al. (2014) most bioinformatician are in one of three groups represented in Figure 2: User, Scientist or Engineer. These groups differ from each other depending on their primary purpose (i.e. analysing biological data or developing an algorithm or software) and skills (i.e. more on the biology or computer science side). It is also important to note how they spend their time working on the computer and/or bench, as well as the time spent working on graphical user interface and/or command line.
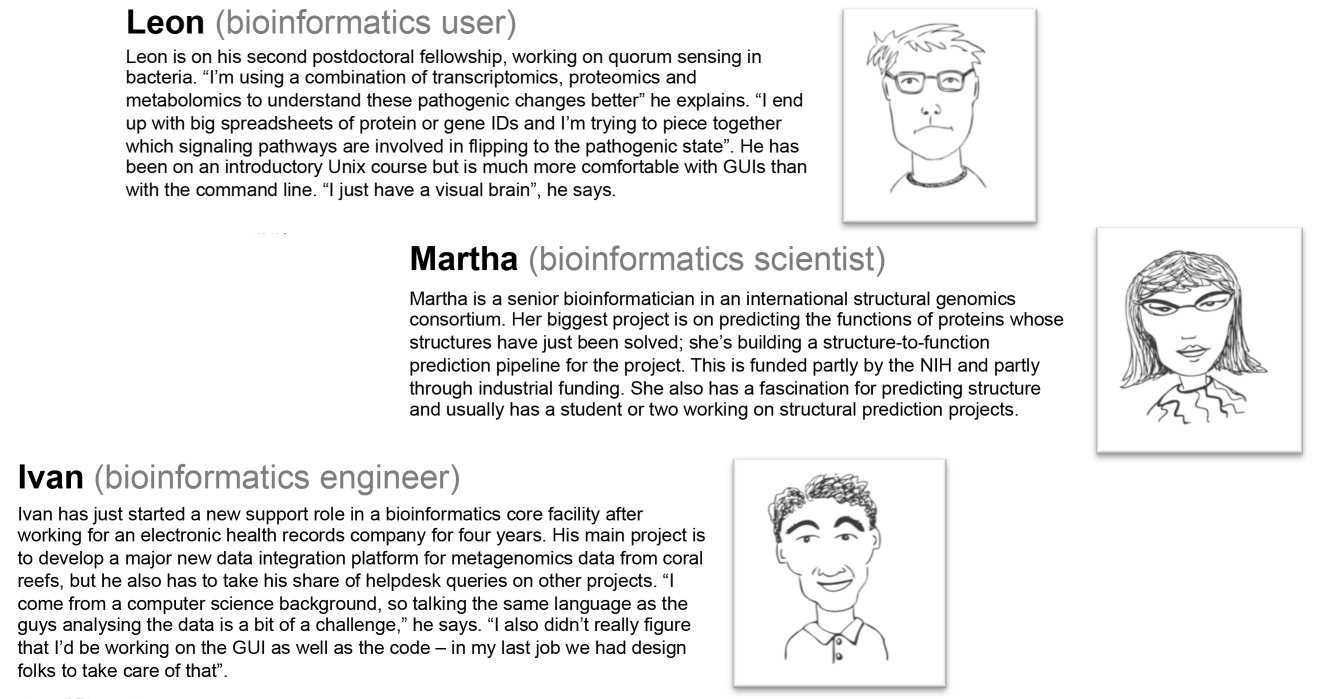
For example, a Bioinformatics User has a specific dataset and wishes to analyse it, obtain results, interpret these results and solve a biological problem. He/she knows how to produce the data and looks for computational tools to generate interpretable results. On the other hand, a Bioinformatics Scientist, although having a good idea of how biological data is generated and interpreted, he/she is more comfortable with mathematical/statistical and computational analysis, including basic programming skills. Finally, a Bioinformatics Engineer is more dedicated to information technology and computer science, and develops or implements software, algorithms and databases that will help the other two groups of bioinformaticians to analyse their biological data.
What are the requirements to become a Bioinformatician?
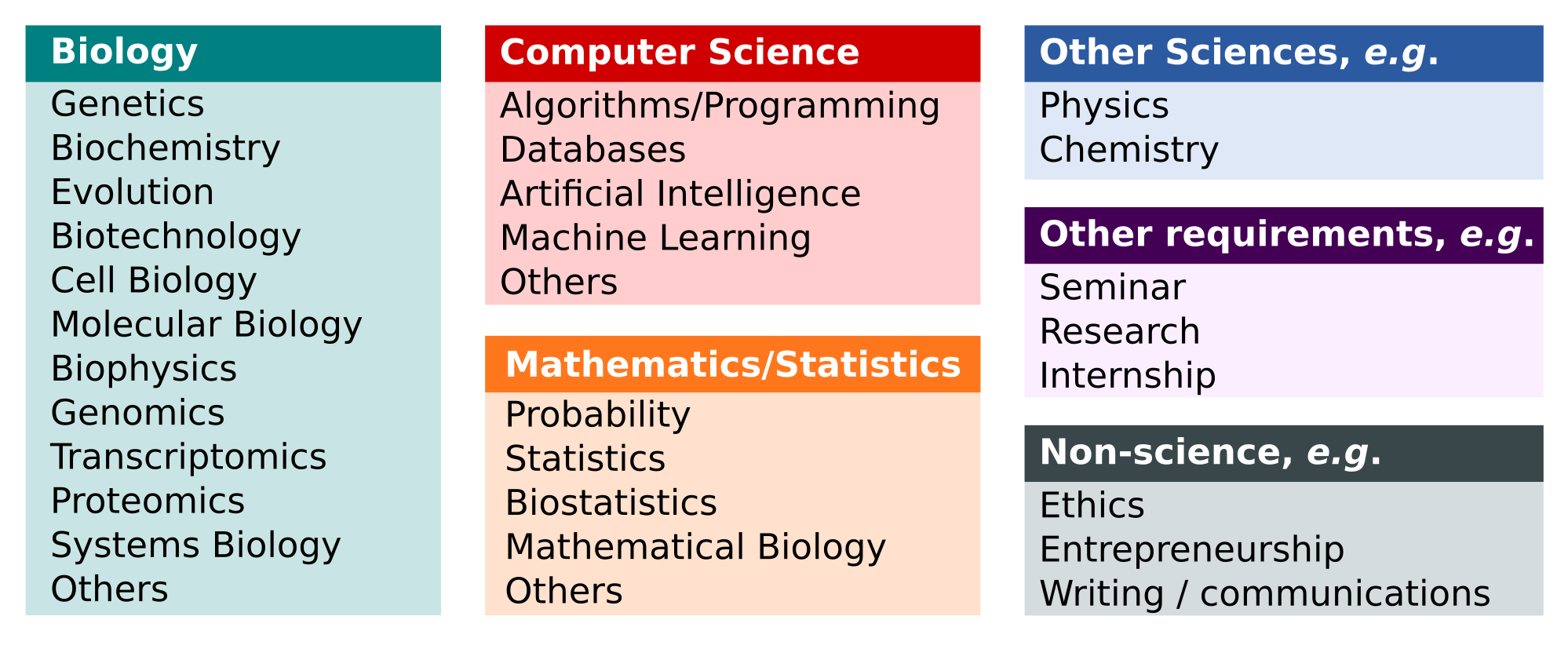
People often enter the field of bioinformatics to learn how to use computer tools to solve biological problems, or to build such tools. That's why a solid background in biology and computer science is extremely useful. Because it is an interdisciplinary area, to become a Bioinformatician you need to learn about different areas of knowledge. For example, from a biological point of view, it is important to know molecular biology techniques, genetics, biochemistry and evolution. On the information technology side, sub-disciplines such as machine learning and data mining are very important as well as programming skills. Figure 3 presents a list of words from a controlled vocabulary that helps to identify requirements for training in bioinformatics. A more detailed view of these requirements can be found in Welch et al. (2014), a publication of the education committee of the International Society for Computational Biology which proposes curricular guidelines and competences in the area.
Where can I find training programs in bioinformatics?
Several countries have masters and doctoral programs that train the student in this area of knowledge. See iscb-degree-certificate-programs for examples of bioinformatics and computational biology programs in the world. In Brazil, besides the UFPR, graduate courses in bioinformatics and computational biology are offered at the UTFPR, UFMG, UFRN, USP, LNCC e Fiocruz. There are also programs that dedicate research lines to bioinformatics (for example, Cellular and Molecular Biology at the UFRGS). For a complete and up-to-date list, see recommended courses by CAPES.
Related videos
Below we have selected some videos that illustrate biological problems that require analysis of large amounts of data, involving the development and/or application of computational tools for scientific discoveries. We have also included some videos that illustrate recurring questions in training a Bioinformatician. If you have more suggestions please contact our program!
This guide is maintained by PPGB under CC BY 4.0, not applied to material in the public domain or any third party content used under another license.
